Introduction
Bacteria live in polymicrobial communities characterized by a great diversity of co-existing species and competition for available resources. Unearthing the molecular mechanisms that shape microbial communities and their dynamics is a major challenge of the post-genomic era. One bacterial strategy for persisting in a specific niche is the production of toxic extracellular factors that interfere with the growth and/or viability of nearby microbes. These ‘bacteriocins’ disrupt the equilibrium among bacterial populations in a phenomenon known as dysbiosis, which can ultimately alter the homeostasis of different host ecosystems, such as humans and plants (Riley and Wertz, 2002). In the 1940s and 1950s, naturally occurring D-amino acids were first identified. Studies revealed that the addition of high concentrations of D-amino acids to bacterial cultures had a powerful effect on morphogenesis and ultimately caused lysis (Fox et al., 1944; Kobayashi et al., 1948; Yaw and Kakavas, 1952; Lark and Lark, 1959; Grula, 1960; Tuttle and Gest, 1960; Bopp, 1965). The effect of D-amino acids on bacterial shape was fundamental in elucidating their mode of action in the peptidoglycan (PG) polymer (also called murein), a specific bacterial structure with a crucial role in preserving bacterial morphology and cell integrity (Caparros et al., 1992). However, these studies recently gained new physiological meaning when it was reported that many taxonomically unrelated bacteria release millimolar concentrations of non-canonical D-amino acids (NCDAAs) into the environment (Lam et al., 2009). In Vibrio cholerae, the causative agent of the diarrheal disease cholera, the periplasmic broad-spectrum racemase BsrV produces mainly D-Met and D-Leu in stationary phase (Lam et al., 2009; Cava et al., 2011). Remarkably, these NCDAAs are incorporated into the PG by replacing the 4th and/or 5th residue of the disaccharide subunit N-acetylglucosamine-(β1-4)-N-acetylmuramic acid-L-Ala-D-Glu-(γ)-di-amino acid-D-Ala-D-Ala, depending on the bacterial species. Cell wall editing via the incorporation of NCDAAs downregulates PG synthesis, thereby permitting synchronization of cell wall metabolism and growth arrest in stationary phase, which preserves cell wall integrity (Lam et al., 2009; Cava et al., 2011). However, incorporation of NCDAAs is also observed in bacteria that do not produce these molecules. ‘Paracrine’ incorporation of NCDAAs causes morphological aberrations and impairs growth in multiple species.
Although virtually all D-amino acids can potentially modify the cell wall, it is unclear why some bacteria produce distinct sets of NCDAAs (Lam et al., 2009; Cava et al., 2011). Here, we report the extracellular production of D-Arg by V. cholerae. Like D-Met and D-Leu, D-Arg is produced by BsrV. However, compared with other D-amino acids, D-Arg exhibits significantly higher inhibitory activity against a wide diversity of bacterial species, suggesting a role of D-Arg as a fitness modulator of bacterial communities in the natural environment. Based on biochemical analyses of PG, microscopy and transposon sequencing, we demonstrate that in contrast to D-Met, which has a major modulatory role in cell wall biosynthesis, the inhibition of growth by D-Arg appears to be cell wall-independent. D-Arginine toxicity is suppressed by mutations in the DnaJ chaperone system and in the Pst phosphate uptake machinery, strongly supporting different roles for NCDAAs in bacterial physiology.
Finally, we analyze the distribution of BsrVs among the Vibrionaceae family and confirm that although highly conserved, production of D-amino acids is not a hallmark of all vibrios. Synteny and racemase activity-based studies confirmed that BsrV orthologues are absent in a number of Vibrionaceae species. In co-cultivation experiments, V. cholerae outcompeted Caulobacter crescentus by D-Arg production. Thus, NCDAAs production by multi-specific racemases might be a strategy employed by vibrios and, potentially, other species to prevail in competitive environments.
Materials and methods
Bacterial strains and culture conditions
Strains are listed in Supplementary Table S1. All strains were grown under optimal conditions (media and temperature) recommended by the DSMZ, ATCC and CECT bacterial collections as indicated in Supplementary Table S1. V. cholerae strains were grown in LB (Luria Bertani broth) media at 37 °C (Dziejman et al., 2002). The Vibrionaceae species were grown in diluted Marine Broth (Difco) at 22 ºC. Defined V. cholerae transposon mutants were taken from an ordered transposon insertion library (Cameron et al., 2008). For stalk length analyses, C. crescentus strains were grown in Hutner base-imidazole-buffered-glucose-glutamate minimal medium supplemented with 10 or 0.03 mm phosphate (Gonin et al., 2000). Agar 1.5% (w/v) was used in solid plates. When required, antibiotics were added to liquid or solid media: Streptomycin (V. cholerae: 200 μg ml−1), Kanamycin (Escherichia coli and V. cholerae: 50 μg ml−1; Agrobacterium tumefaciens: 300 μg ml−1) and Ampicillin (E. coli: 100 μg ml−1). Media was supplemented with L- or D-amino acid at the indicated concentration when needed.
For growth curves, at least three replicates per strain and condition tested were grown in two independent experiments in 200 μl medium in a 96-well plate inoculated 1:1000 from exponentially growing precultures. Optical density at 600 nm (OD600) was monitored using an Eon Biotek plate reader (Biotek, Winooski, VT, USA) at 10 min intervals and the optimal growth temperature.
Viability assays
Overnight cultures were normalized to 1 unit of optical density at 600 nm (OD600) and subjected to serial 10-fold dilution. Five-microliter drops of the 100 through 10−7 dilutions were then spotted onto the indicated agar plates and incubated at the appropriate temperature for 24–48 h prior to image acquisition.
Bacterial competition assay
A NCDAA producer (V. cholerae) and a NCDAA-sensitive bacteria (C. crescentus) were inoculated in compartments A and B respectively of a Stericup system separated by a 0.22 μm membrane. Both compartments were filled with peptone yeast extract (PYE) media, for optimal growth of C. crescentus, and supplemented or not (control) with 5 mm L-Arg. Compartment A was inoculated with V. cholerae wild-type (lacZ+), ΔbsrV mutant (lacZ−), a 1:1 mix of both or none (-). Compartment B was inoculated with C. crescentus. Devices were incubated at 28 °C with mild-agitation (100 rpm). The 0.22 μm filter avoids bacterial contact while allowing free diffusion of small molecules such as amino acids. Both viability and D-amino acid concentration were measured at different time points (0, 24, 48 and 72 h). Viability was assessed by CFU ml−1 counting on LB (V. cholerae) or PYE (C. crescentus) agar plates. The ratio WT/ΔbsrV in V. cholerae mix was assessed by plating on LB plates containing X-gal 40 μg ml−1, to distinguish between WT (lacZ+, blue) and ΔbsrV (lacZ−, white) colonies. D-amino acid concentration was determined using a D-amino acid oxidase assay (see below). The competition assay was performed twice in triplicates.
PG analysis
PG was isolated and analyzed following previously described methods (Desmarais et al., 2013; Alvarez et al., 2016). In brief, PG sacculi were prepared by boiling bacterial cells in SDS 5%. After SDS removal by ultracentrifugation, the insoluble material was further digested with muramidase (Cellosyl). Soluble muropeptides were separated by liquid chromatography (high-performance liquid chromatography and/or ultra high-pressure liquid chromatography) and identified by MALDI-TOF and electrospray-ion trap MS. A detailed protocol is described in Extended Experimental Procedures.
Total PG amount was determined by quantification of total intensities of the PG profile from three biological replicas normalized to the same OD600 ml−1. Quantification of relative abundances of muropeptides was calculated as the relative area of the corresponding peak compared to the total area of the normalized chromatogram.
D-amino acid analysis and quantification
D-amino acids in the supernatant were derivatized with Marfey’s reagent (Bhushan and Bruckner, 2004) and analyzed by high-performance liquid chromatography as described previously (Espaillat et al., 2014). Total D-amino acid concentration was determined by D-amino acid oxidase assay coupled to peroxidase and 2,3 diaminophenazine (Espaillat et al., 2014). Detailed protocols are described in Extended Experimental Procedures.
Microscopy
Bacteria were immobilized on LB or PYE pads containing 1% agarose. Phase contrast microscopy was performed using a Zeiss Axio Imager.Z2 microscope (Zeiss, Oberkochen, Germany) equipped with a Plan-Apochromat × 63 phase contrast objective lens and an ORCA-Flash 4.0 LT digital CMOS camera (Hamamatsu Photonics, Shizuoka, Japan), using the Zeiss Zen Blue software. Image analysis and processing were performed using Fiji and the MicrobeJ plugin (Schindelin et al., 2012; Ducret et al., 2016). Sphere-inducing activity of D-amino acids was analyzed by cell shape analysis of 380–480 cells per condition.
Transposon insertion sequencing
For the identification of essential genes, transposon insertion sequencing (TnSeq) was performed as described previously (Chao et al., 2013). In brief, 3 × 105–4 × 105 transposon mutants were generated for each library by conjugation of V. cholerae with E. coli SM10λPIR carrying pSC189 (transposon donor plasmid) (Chiang and Rubin, 2002). Mutant libraries were selected under three conditions (control, 5 mm D-Arg and 5 mm D-Met) and pooled genomic DNA fragments were analyzed using a MiSeq sequencer (Illumina, San Diego, CA, USA). Insertion sites were identified and significance was determined using ConArtist simulation-based normalization as described (Chao et al., 2013; Pritchard et al., 2014).
Suppressor mutants
To obtain suppressor mutants, C. crescentus was grown in 2 ml PYE supplemented with 0.5 mm either D-Arg or D-Met. Cultures were grown for 24 h at 28 °C and reinoculated in fresh media containing the corresponding D-amino acid during 12 days (80 generations). After this preadaptation process, 0.1 ml aliquots (~1 × 108 cells) were inoculated onto PYE agar plates containing 1–10 mm D-amino acid. Plates were incubated at 28 °C until suppressor mutant colonies arose. For confirmation of the resistance, the selected derivatives were grown in presence and absence of the corresponding D-amino acid during several generations as described above, prior to viability testing in presence of D-amino acid.
Suppressor mutants of A. tumefaciens were obtained likewise by preadaptation in LB 0.5 mm D-Arg media during 12 days (80 generations) and selection of resistant derivatives on 10 mm D-Arg containing plates.
Whole-genome sequencing and single-nucleotide polymorphism analysis
Genomic DNA samples from suppressor mutants and parental strain were prepared. Indexed paired-end libraries were constructed and sequenced in a MiSeq sequencer (Illumina), following the manufacturer’s instructions. The sequences were analyzed using the Galaxy server tools (https://usegalaxy.org/, (Afgan et al., 2016)) for identification of variants (single-nucleotide polymorphisms). A detailed protocol is described in Extended Experimental Procedures.
Reconstruction of suppressor mutant pstBM108T in A. tumefaciens
For reconstruction of the point mutation pstBM108T in A. tumefaciens, a 1000 bp fragment containing the mutation site was amplified from purified genomic DNA with primers FCP2041 (5′-AAAAAAGCTTTCCGACAACGCCGATGGA-3′) and FCP2044 (5′-AAAAAAGCTTGAGGCACCAGTCTTGTTC-3′) and cloned into plasmid pNPTS139 (Fischer et al., 2002). E. coli DH5α λPIR was used in the cloning step and the resulting plasmid pNPTS139pstBM108T was verified by sequencing.
Nucleotide substitution in A. tumefaciens pstB gene (Atu0423) was performed following an established allelic-replacement protocol (Morton and Fuqua, 2012). In short, exconjugants obtained by conjugation with E. coli S17-1 λPIR cells as a donor of pNPTS139pstBM108T were selected on ATGN plates containing Kanamycin 300 μg/ml. Exconjugants were grown in ATGN medium overnight and then plated on ATSN plates containing 5% (w/v) sucrose. Colonies sensitive to kanamycin were streak-purified twice on ATSN plates and checked by sequencing.
Results
V. cholerae releases high concentrations of D-arginine to the extracellular medium
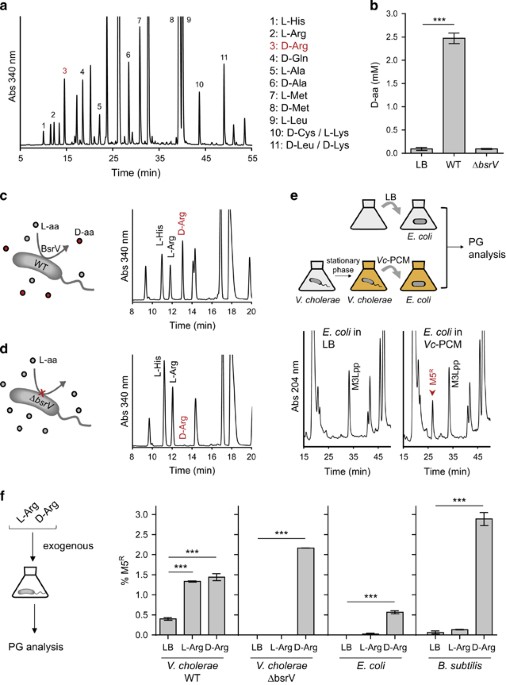
An exhaustive chemical analysis of the stationary-phase extracellular medium of V. cholerae revealed that this bacterium releases a greater diversity of D-amino acids than previously reported (Figure 1a). In addition to D-Met and D-Leu (Lam et al., 2009), we detected D-Gln, D-Cys, D-Lys and D-Arg, whose identities were confirmed by supplementing the samples with pure D-amino acid standards (for example, D-Arg, Supplementary Figure S1a). Using a D-amino acid oxidase activity-based assay, we determined that V. cholerae releases approximately 2.5 mm of D-amino acids (Figure 1b), including ~0.7 mm D-Arg.
Figure 1 D-arginine is released to the extracellular medium. (a) HPLC profile of Marfey’s derivatized preconditioned medium (PCM) from stationary-phase V. cholerae cultures for quantification of chiral amino acids. L- and D-forms were identified by comparison to standards. (b) Total D-amino acid content in wild-type and ΔbsrV mutant V. cholerae cultures, determined by D-amino acid oxidase assay. P-value
D-arginine is released to the extracellular medium. (a) HPLC profile of Marfey’s derivatized preconditioned medium (PCM) from stationary-phase V. cholerae cultures for quantification of chiral amino acids. L- and D-forms were identified by comparison to standards. (b) Total D-amino acid content in wild-type and ΔbsrV mutant V. cholerae cultures, determined by D-amino acid oxidase assay. P-value