Main
Most bacteria can sense and respond to a wide variety of signals to thrive in changing environments. One such adaptive response is chemotaxis, by which motile bacteria monitor changes in local concentrations of certain substances and initiate a signalling cascade that, over time, biases bacterial movement towards favourable conditions (for example, nutrients) or away from toxic compounds1.
Albeit species-specific components exist, the core of the chemotaxis signalling pathway consists of chemoreceptors, known as methyl-accepting chemotaxis proteins (MCPs), the adaptor protein CheW, the histidine kinase CheA and the response regulator CheY. In some species such as Escherichia coli, the MCPs form a stable ternary sensory complex with CheW and CheA that typically clusters at cell poles. Upon ligand binding or other environmental perturbations, the MCPs change conformation and modulate CheA kinase activity with the help of CheW. CheA in turn phosphorylates CheY, thereby allowing it to bind to the flagellar motor to induce clockwise rotation and, thus, cell tumbling2,3. To ensure a rapid response to chemotactic stimuli, the phosphorylation of CheY can be controlled by a specific phosphatase4. Although the chemotaxis signalling core components are highly conserved among bacteria and archaea, the number and type of chemoreceptors are highly variable between species and their specificities remain largely uncharacterized5,6,7.
Compared to E. coli, which has a single chemosensory system and 5 MCPs, the facultative pathogen Vibrio cholerae has a very sophisticated chemotaxis system organized into three sets of chemosensory pathways (Che systems I/F9, II/F6 and III/F7) and at least 45 chemoreceptors8. Thus far, only system II/F6 has been demonstrated to control motility9, while the function of the others remains unclear. The high number of MCPs has been suggested to reflect the complexity of V. cholerae’s life cycle10; however, except for a handful of input signals (for example, l-amino acids11, taurine12, oxygen13) there is almost no information about the ligand specificity of most MCPs.
V. cholerae releases millimolar concentrations of diverse d-amino acid (DAAs) into the environment14,15 (Fig. 1a). DAAs are produced from their l-amino acid (LAA) counterparts by the broad-spectrum racemase BsrV, which is conserved in many bacterial species14. These molecules regulate diverse cellular processes (for example, cell wall biogenesis16,17, biofilm integrity18,19,20,21, spore germination22 or bacteria–bacteria interactions15) but overall, the physiological role of extracellular DAAs is mainly dependent on each specific d-amino acid type and bacterial species. In V. cholerae, it is known that d-Met and d-Leu modulate cell wall biogenesis, but the function of other DAAs is unclear.
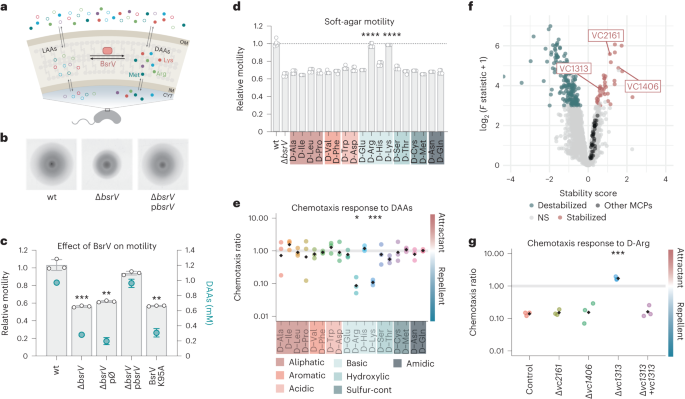
Fig. 1: d-Arg and d-Lys signal a chemorepellent response in V. cholerae through the putative MCP VC1313. a, Schematic representation of DAA racemization. The most abundant amino acids produced are shown (open circles, l-amino acids; filled circles, d-amino acids). OM, outer membrane; IM, inner membrane; CYT, cytoplasm. b, Representative images of V. cholerae wild type (wt) and ΔbsrV mutant expanding in 0.3% soft agar. Images are representative of experiments repeated at least three times. c, Motility and d-amino acid production analysis of BsrV derivatives; grey bars indicate relative motility compared to wt or ΔbsrV knockout mutant, catalytic site mutant (BsrV K95A) and the complemented strain harbouring bsrV under an aTc inducible promoter (pbsrV); green dots represent the amount of secreted d-amino acids. pØ, empty plasmid. Error bars represent mean ± s.d. of 3 biologically independent replicates. Significant differences (paired t-test) are indicated by **P
a, Schematic representation of DAA racemization. The most abundant amino acids produced are shown (open circles, l-amino acids; filled circles, d-amino acids). OM, outer membrane; IM, inner membrane; CYT, cytoplasm. b, Representative images of V. cholerae wild type (wt) and ΔbsrV mutant expanding in 0.3% soft agar. Images are representative of experiments repeated at least three times. c, Motility and d-amino acid production analysis of BsrV derivatives; grey bars indicate relative motility compared to wt or ΔbsrV knockout mutant, catalytic site mutant (BsrV K95A) and the complemented strain harbouring bsrV under an aTc inducible promoter (pbsrV); green dots represent the amount of secreted d-amino acids. pØ, empty plasmid. Error bars represent mean ± s.d. of 3 biologically independent replicates. Significant differences (paired t-test) are indicated by **P